Loss of heterozygosity in the short tandem repeat (STR) loci found in tumor DNA of ALL patients as a factor predicting poor outcome
National Research Center for Hematology, Moscow, Russian Federation (Russia)
Genetic instability of tumor cells can lead to a change in the profile of short tandem repeats (STR) of the tumor relative to the STR profile of healthy cells of the patient. Loss of heterozygosity or allelic imbalance in STR loci confirm the chromosomal aberrations revealed by standard cytogenetic analysis in chromosomes containing this STR locus: deletions, monosomies, duplications, and the appearance of isochromosomes. However, in some patients, the loss of heterozygosity (LOH) is also observed with a normal tumor karyotype. Our objective was to evaluate correlations between the loss of heterozygosity in blast cells of de novo diagnosed ALL patients and chromosomal aberrations revealed by standard cytogenetic analysis, as well as to verify hidden abnormalities in the case of discrepancies between cytogenetic data and molecular analysis of the STR profile of the tumor.
Due to some restrictions caused by COVID quarantine the control points schedule was insignificantly delayed.
A study of a cohort of patients with ALL for the loss of STR-markers heterozygosity was performed in two periods: May-June and August-September 2020. Chromosomal microarray (CMA) was done in September-October 2020. The CMA results of LOH boundaries and genes involved in the field of uniparental disomy, deletions and duplications were analyzed and statistical evaluation of the loss of heterozygosity impact on overall and event-free survival was made in September-October 2020. Analysis of STR profiles of tumor cells was performed in 88 patients with de novo diagnosed Ph-negative ALL undergoing treatment according to the RALL-2016 protocol at the National Research Center of Hematology (Moscow, Russia).
The patients were 18-55 years old, 53 men and 35 women. DNA was isolated from bone marrow samples taken from patients at diagnosis. Control DNA samples were taken from the blood of patients in complete remission and/or from the buccal epithelium. STR profiles for each pair of tumor/control samples were obtained by multiplex PCR using the COrDIS Plus kit (Gordiz, Moscow), followed by fragment analysis of PCR products on an ABI 3130 genetic analyzer (Thermofisher Scientific, USA). Chromosomal microarray (CMA) of DNA with a detected loss of heterozygosity in STR loci was performed at the Genomed labs (Moscow, Russia) molecular pathology laboratory using the Genoscan 3000 system.
Results
Normal cytogenetic tumor karyotype was established for 37 patients, and abnormal karyotypes were found in 51 cases. When analyzing the STR profiles of DNA from tumor and healthy cells of each patient, we found a loss of signal from one of a pair of alleles at heterozygous STR loci in twenty-one patients (24%). Seven of them had a normal tumor karyotype. Also, in the case of an abnormal STR karyotype, the loci with loss of heterozygosity did not always belong to aberrant chromosomes. Of 14 people with an abnormal tumor karyotype, for 8 the analysis of STR-profile revealed an allelic imbalance in loci, in which the CMA method was confirmed additional chromosomal aberrations not detected by cytogenetic analysis. And for 6 people LOH in STR loci confirmed by CMA coincided with cytogenetic analysis data. In this way, analysis of STR profiles revealed additional disorders in 15 out of 88 people(17%). In these 15 patients with CMA, aberrations up to 190 million base pairs (more than 1000 genes) were found. In the majority of patients with changes in the STR-profile on CMA, additional chromosomal abnormalities were found that were not identified by cytogenetic analysis and analysis of STR-profile.
One of these abnormalities was uniparental disomy of the shoulder or the entire chromosome, which was cytogenetically normal, but carried an STR locus with loss of heterozygosity. We issued the frequency of LOH and chromosomal aberrations found by CMA. The most frequent was LOH at 12p about 38% (8 from 21 patients) identified by CMA as a deletion in 25%, duplication in 6%, and uniparental disomy in 6%. Patients with the LOH at the short arm of chromosome 12 (12p) are excluded from the risk group because 12p LOH is not associated with worse outcome according to some publications [Takeuchi et al Leukemia 2003, Kawashima-Goto et al Int J Hematol. 2015].
LOH at 9p was also most frequent in ALL group. It was confirmed by CMA as a deletion in 38% (8 from 21 patients) and uniparental disomy in 19% (4 from 21 patients). *Clinical case: For one patient with a normal karyotype and STR-marker LOH, chromosomal microarray showed the microdeletion at 9p21.3 (21656682_22304230) affected the region of uniparental disomy at 9p (24.3-13.3). As a result of two of these chromosomal aberrations leading to a homozygous form of deletion, a cluster of MTAP, CDKN2A-AS1, CDKN2A, CDKN2B-AS1, CDKN2B genes containing three oncosuppressor genes involved in the regulation of antiproliferative and proapoptotic activities of Rb1 and p53, was completely absent from the tumor cell genome. The patient died of the disease. LOH 9p seems the most unfavorable marker, but it occurs in combination with other LOH that complicates the evaluation of LOH 9p as an independent risk factor.
Statistical Analysis
LOH is a possible independent cofounder for Relapse Free Survival and Overall Survival of ALL patients. The null hypothesis – the loss of heterozygosity isn`t associated with the worst relapse-free and overall survival. Multivariate survival analysis was used to assess the independent impact of the new risk factor against the background of the other standard factors for this cohort of patients. Based on the preliminary obtained data, we expected the risk group according to the criteria for loss of heterozygosity would be 15-20% of the entire cohort. But in our project LOH group was 24% (21 patients from 88). OS and EFS analysis had shown, that the LOH presents a significant risk factor for dismal prognosis (Hazard Ratio= 4.1 (ci95 1.1-15.6)) (Fig.1)
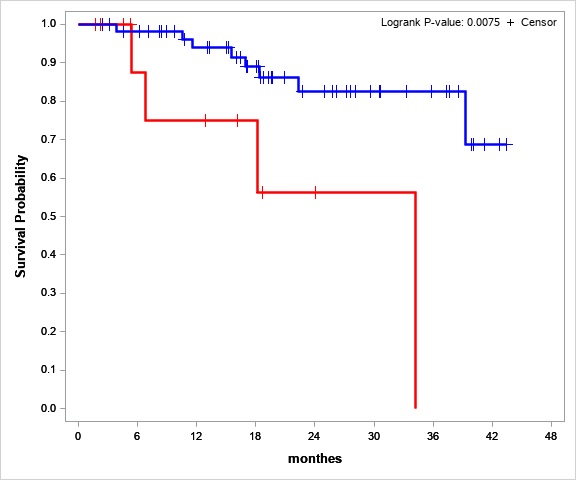
Figure №1 Overall survival estimates for stable STR-profile (blue line) and LOH (red line) patients
We also tried to evaluate LOH 12p as a risk factor. Overall survival in the LOH 12p group was 100%. LOH 12p seems to the marker of preferable prognosis, but the group with LOH 12p only (without any other LOH) is too small (three people) for statistical significance evaluation.
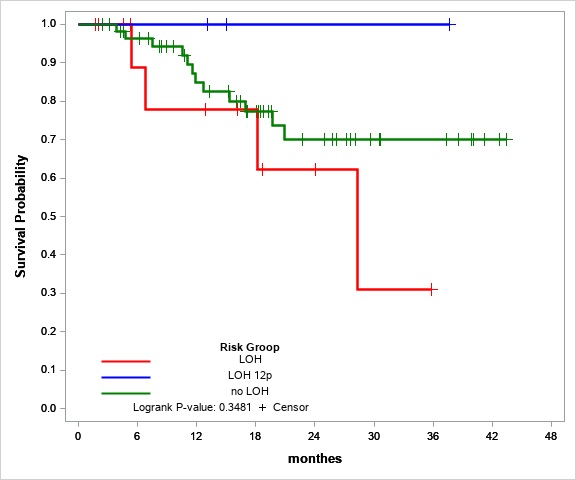
Figure №2 Overall survival estimates for stable STR-profile (green line), LOH (red line) and LOH 12p (blue line) patients
Conclusion
Analysis of STR-profile in the tumor cells confirms and supplements the data of cytogenetic analysis, and also reveals an abnormal karyotype in the absence of mitoses in the sample. Moreover, LOH detection in cases of normal tumor karyotype reveals the phenomenon of uniparental disomy, hidden chromosomal aberration, which is often the cause for the transition of oncogenic mutations to homozygous form, thus aggravating the prognosis of the disease. The study of LOH 12p as favorable and LOH 9p as worst prognostic factors requires research on an expanded sample of patients.
The results were presented at the Society of Hematologic Oncology 2020 annual meeting on September 9-12, 2020 (poster presentation), prepared for III Congress of hematologists of Russia (canceled) , presented at XIV Raisa Gorbacheva Memorial Symposium “Hematopoietic Stem Cell Transplantation. Gene and Cellular Therapy” and at XXVII International Scientific Conference of Students, Postgraduates and Young Scientists “Lomonosov” 10 – 27 Nov 2020 (poster and oral presentation, https://cloud.mail.ru/stock/mUcEsH1wDX4aaPGE6ACymvHL). The original articles are being prepared for publication, but due to force majeure (restrictions associated with covid-19), everything has been delayed.

