Loss of Heterozygosity in the Short Tandem Repeat (STR) Loci Found in Tumor DNA of De Novo Diagnosed ALL Patients As a Factor Predicting Poor Outcome: Interim Results Of The Study
This study was supported by RakFond, Grant No. 5/2019
KEY WORDS: Loss of heterozygosity (LOH), de novo diagnosed ALL, chromosome aberration, hidden chromosomal abnormalities, uniparental disomy
Introduction
Acute lymphoblastic leukemia (ALL) is a heterogeneous group of the blood clonal diseases characterized by the transformation of precursors of B or T cells into leukemic cells.
ALL accounts for less than a third of all acute leukemias in adults, which is diagnosed most often in young people (median age is 30-35 years old). For the therapy of acute lymphoblastic leukemia to be successful, it is important to have the most complete cytogenetic and molecular characteristics of the tumor.
Genetic instability in tumor cells may lead to the changes in STR profiles of the tumor in relation to the STR profile of the patient’s normal cells (Fig. 1).
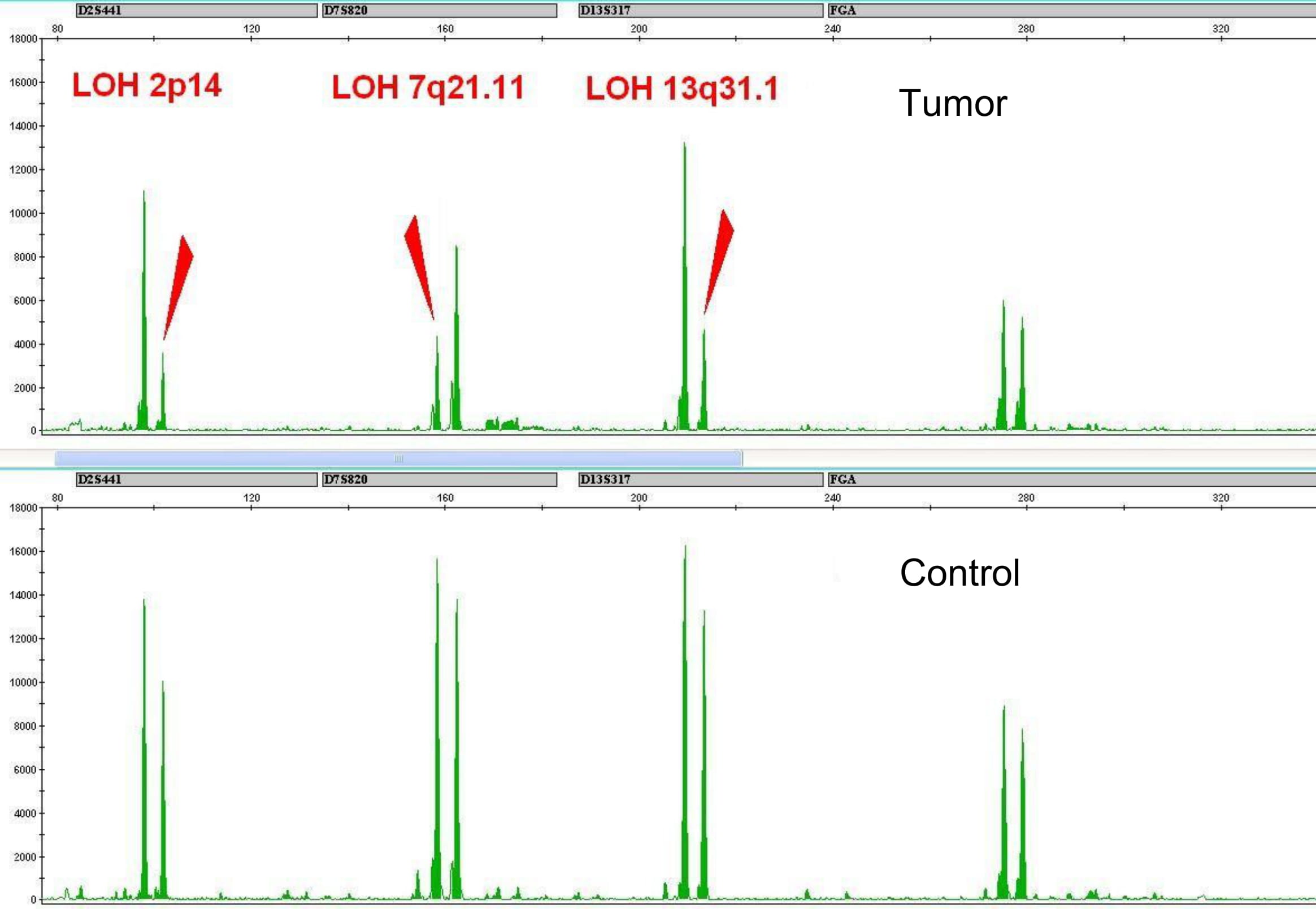
*STR (short tandem repeats) are short segments (loci) of DNA consisting of tandem repeat monomers of length less than 9 base pairs and forming the fields of less than 1 thousand base pairs.
Loss of heterozygosity or allelic imbalance in STR loci confirm the chromosomal aberrations containing this STR locus by standard cytogenetic analysis: deletions, monosomy, duplications, formation of isochromosomes.
However, in some patients, the loss of heterozygosity is also observed with a normal karyotype of the tumor. The reason is chromosomal aberrations that are not detected by standard cytogenetic analysis and FISH probes. This is a uniparental disomy (UPD) or the loss of heterozygosity with the preserved number of DNA copies (Fig. 2). We assume that the patients with these chromosomal abnormalities are at high risk and should receive appropriate treatment.
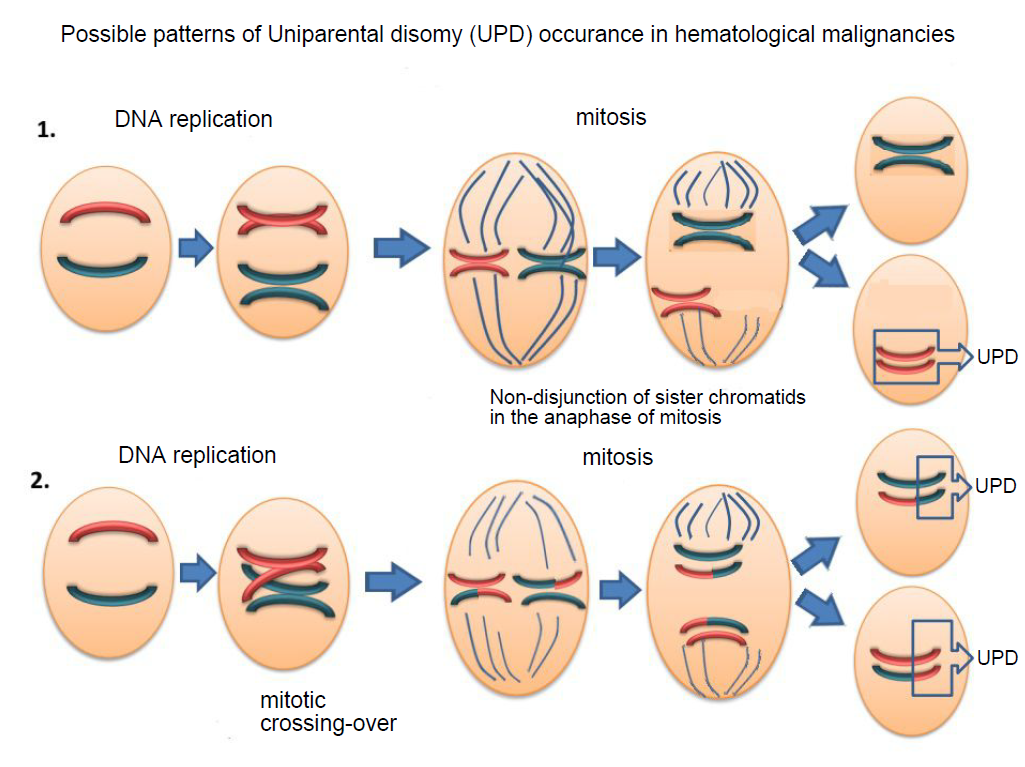
Fig.2 Reasons of UPD occurrence in tumor cells.
1. Non-disjunction of sister chromatids in the anaphase of mitosis.
2. Mitotic recombination. During the S-phase of mitosis and after the chromosomes’ duplication, paired chromatids of homologous chromosomes may swap segments of DNA. As the result of random chromatid segregation, recombinant and normal chromatids may get into one sister-cell, which leads to the loss of heterozygosity at the recombinant locus.
Objectives. To investigate the relationship between the loss of heterozygosity in blast cells of patients with newly diagnosed ALL and the chromosomal aberrations detected by standard cytogenetic analysis and to verify the hidden abnormalities if there are inconsistencies between cytogenetic data and molecular analysis of the STR profile of the tumor. To evaluate the overall survival of patients in the assumed high-risk group compared to other patients.
Materials and methods. Analysis of the STR profile of tumor cells’ DNA was carried out for 88 de novo diagnosed Ph-negative ALL patients undergoing treatment according to the “ALL-2016” regimen at the National Research Center for Hematology (Moscow, Russia). 53 male and 35 female patients aged 18-55, all at intermediate risk according to the applicable clinical and diagnostic criteria.
DNA was isolated from the bone marrow samples taken from patients at diagnosing. Control DNA samples were taken from the blood of patients in complete remission and/or from the buccal epithelium (buccal swab).
STR profiles of each pair of DNA samples (tumor/control) were obtained by multiplex polymerase chain reaction with COrDIS Plus multiplex kit for amplification of 19 polymorphic STR markers and amelogenin loci (Gordiz Ltd, Russia) with further fragment analysis of PCR products performed with genetic ABI 3130 (Thermofisher Scientific, USA).
Chromosomal microarray analysis (CMA) of DNA with detected loss of heterozygosity in STR loci was performed at the Genomed molecular pathology laboratory using the Genoscan 3000 system.
Results of the discussion. Normal and abnormal cytogenetic tumor karyotypes were identified in 37 and 51 patients, respectively. Analyzing the patients’ STR profiles of DNA from tumor and normal cells, we observed a loss of signal from one pair of alleles in heterozygous STR loci in twenty patients (23%).
Normal karyotype of the tumor was identified in seven of these patients. Moreover, even in case of the abnormal karyotype the STR loci with the loss of heterozygosity were not always attributed to the aberrant chromosomes.
Chromosomal microarray analysis indicated the presence of uniparental disomy of the entire chromosome or its arm, which was normal according to cytogenetic analysis data, but carrying the STR locus with the loss of heterozygosity.
Thus, one patient with a normal karyotype had a 9p21.3(21656682_22304230)x0 microdeletion which occured in the 9p (24.3-13.3) region of the uniformed disomy, and as the result, the cluster of MTAP, CDKN2A-AS1, CDKN2A, CDKN2B-AS1, CDKN2B genes having three onco-suppressor genes, which products are involved in the regulation of the antiproliferative and proapoptotic activities of the Rb1 and p53 genes, was not presented in the tumor genome at all (Fig. 3).
This patient died from disease progression due to the resistance to treatment.
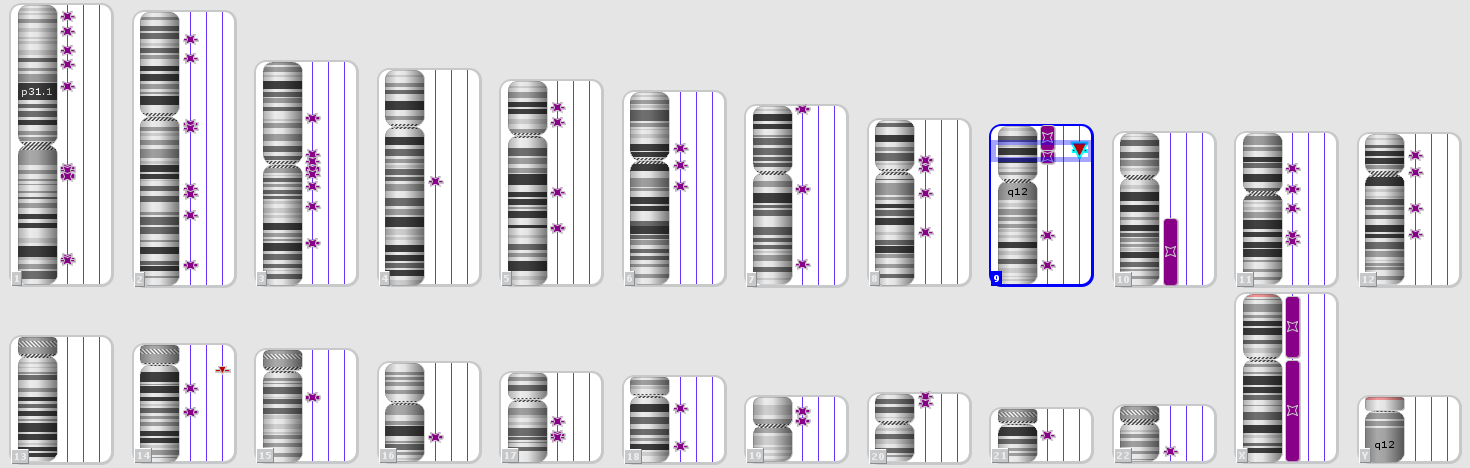
Fig. 3. Karyotype according to the Chromosomal microarray analysis of a patient with the loss of heterozygosity and normal karyotype according to the standard cytogenetic analysis. Detailed image of chromosome 9 with the deletion 9p21.3(21656682_22304230).
The preliminary results of overall survival assessment showed that the loss of heterozygosity is a significant factor associated with poor outcome. The probability of an adverse event (death) in the group with the loss of heterozygosity is 4.11 (CI95 1.08-15.63) times higher (Fig. 4).
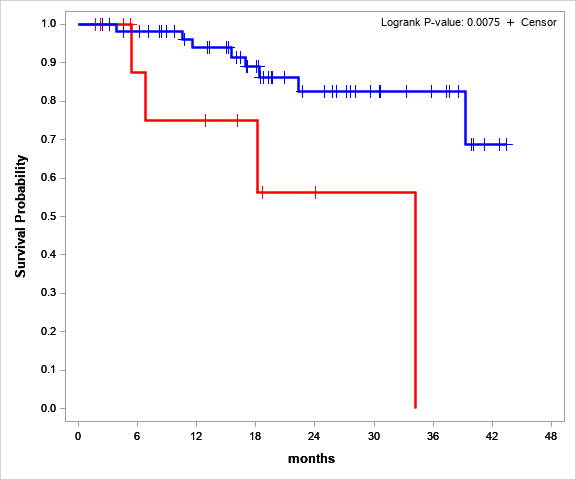
Fig.4 Overall survival of patients at assumed high risk (red) and patients in the control group (blue), P value = 0,0075
Additionally, the analysis of clinical and laboratory indicators in patients at the onset of the disease demonstrated that the level of platelets in the risk-group is significantly lower than in the control group. We do not have an explanation for this yet.
Conclusion. Analysis of the STR profile of the tumor confirms and complements the results obtained from the cytogenetic analysis, as well as reveals an abnormal karyotype in case of the absence of mitosis in the cells.
Moreover, the loss of heterozygosity with normal karyotype reveals the phenomenon of uniparental disomy, which is often the cause of the transition of oncogenic somatic mutations to the homozygous form. According to our data, the identified loss of heterozygosity even with a normal karyotype of tumor cells significantly aggravates the prognosis of the disease.
This study was supported by RakFond, Grant No. 5/2019
Multivariate survival analysis was used to assess the independent impact of LOH as a risk factor. Overall survival (OS) was chosen as the primary endpoint. Patients with LOH on the short arm of chromosome 12 (12pLOH) were excluded from the risk group, as according to several publications 12p LOH is not associated with the worse outcome [1,2].
- Takeuchi, S., Tsukasaki, K., Bartram, C. et al. Long-term study of the clinical significance of loss of heterozygosity in childhood acute lymphoblastic leukemia. Leukemia 17, 149–154 (2003). https://doi.org/10.1038/sj.leu.2402727
- Kawashima-Goto S, Imamura T, Seki M, et al. Identification of a homozygous JAK3 V674A mutation caused by acquired uniparental disomy in a relapsed early T-cell precursor ALL patient. Int J Hematol. 2015;101(4):411-416. doi:10.1007/s12185-014-1711-y
Loss of heterozygosity (LOH) in the tumor DNA is not covered by the standard list of molecular tests and mostly detected accidentally when monitoring chimerism (the ratio of genetically different donor and recipient cells) using the analysis of STR loci in patients after q bone marrow transplantation in the relapse (recurrence) of the disease.
It was important for us to perform this specific study on a large setting of de novo diagnosed patients with a common diagnosis and treatment in order to prove that the loss of heterozygosity in STR loci reflects serious hidden chromosomal events and has an impact on the outcome of the disease. With the support of RakFond we were able to verify these aberrations using the method of chromosomal microarray analysis and found that the additional major changes occurred in the genome of tumor cells in all patients with identified loss of heterozygosity, both with normal and abnormal karyotypes, in addition to the events found with the standard cytogenetic analysis and FISH.
After evaluating the overall and event-free survival we were convinced that the loss of heterozygosity revealed by STR profiling of the tumor DNA in relation to the DNA from normal cells is an independent factor of poor prognosis. STR profiling is an affordable and simple method for the integral assessment of changes in the tumor genome. We hope that, based on the results of our study, this method will gradually be adopted into the routine practice of molecular studies in acute leukemia in addition to standard cytogenetics or even as the main method for assessing the DNA from archival materials (for example, archival slides with bone marrow smears) when a cytogenetic analysis is simply impossible to perform.
In line with the undoubted practical significance of the obtained results, this study has also helped us to expand our understanding of the nature of cell malignancy and the role of uniparental disomy in this process. We observed it with our own eyes, and we are very grateful to RakFond for financing the reagents for the chromosome microarray analysis.
We have studied all available data from the patients receiving treatment in accordance with the randomized “ALL-2016” regimen at the National Research Center for Hematology (Moscow, Russia) and conclude that the loss of heterozygosity is statistically significant as a factor of the poor outcome.
We have yet to compare the data of chromosomal microarray analysis, STR profiling, and standard cytogenetic analysis to determine the range of possible chromosomal aberrations with the loss of heterozygosity in each STR marker of the Cordis + panel. We also plan to continue our studies after the grant expires and expand the study cohort of patients for identification of STR markers, where the loss of heterozygosity has the greatest impact on the outcome of the disease.

